Questions & Challenges
Microorganisms are the foundation of the Earth’s biosphere, and play integral and unique roles in ecosystem functions and biogeochemical cycling of carbon (C), nitrogen (N), sulfur (S), phosphorus (P), and various metals. They are the most diverse group of life presently known, inhabiting almost every imaginable environment on Earth. But they never live alone. Instead, they grow together to form complex communities and these communities are always undergoing dynamic structural change over space and time. Understanding the structure, functions, interactions, and population changes of microbial communities over time and space is critical for many aspects of our lives, including science discovery, biotechnology development, sustainable agriculture, energy security, environmental protection, and human health. For instance, sudden, dramatic changes of microbial communities from human epidemics, plant or animal diseases, or atmospheric alternations due to global climate change could represent a disaster to us or our environment. But, how do we know “who is there and what are they doing with whom and when” in these ecosystems?
Detection, identification, characterization, and quantification of microbial communities face several grand challenges. First, microbial communities are extremely diverse in familiar environments such as soil, water, food, and within our own bodies. One gram of a typical soil contains more than 5,000 (up to 40,000) microbial species. Characterizing such a vast diversity and understanding the mechanisms shaping it presents numerous obstacles. Second, the majority of these microorganisms (>99%) have not yet been cultured, which presents enormous difficulty for microbiologists to study microbial identity and community structure, activity, and dynamics. In addition, although microorganisms control, at least to some degree, various ecosystem processes, in most cases, we do not know what they are doing. Establishing mechanistic linkages between microbial diversity and ecosystem functioning is even more difficult. To address these challenges, revolutionary high-throughput detection technologies for analyzing microbial communities are needed.
To assess microbial community composition, structure, functions, and dynamics in natural settings, microbial detection tools must be:
- simple, rapid, and robust;
- specific and sensitive with a broad comprehensive coverage of target microorganisms;
- quantitative and accurate with wide dynamic ranges;
- capable of detecting functions with high resolution;
- capable of high throughput and in parallel analysis;
- capable of making reliable comparisons across different sites, experiments, laboratories and/or time periods;
- cost-effective.
No such accurate, comprehensive, high-throughput, and cost-effective approaches have been developed thus far to characterize microbial community functional structure and activity. Traditional culturing techniques have proven difficult and ultimately, provide an extremely limited view of microbial diversity and functions. Although conventional nucleic acid detection approaches, such as 16S rRNA gene-based cloning methods, denaturing gradient gel electrophoresis (DGGE), terminal-restriction fragment length polymorphism (T-RFLP), quantitative PCR, and in situ hybridization remain vital to studies of microbial communities, they do not easily meet these requirements. These individual gene-based molecular approaches are slow, laborious, or low-throughput, have low resolution, are not quantitative and/or provide very limited functional information. Thus, they are not suitable for high throughput and comprehensive functional characterization of microbial communities at the whole community level.
GeoChip as a Solution

To address those challenges, GeoChips have been developed, which have unique features to answer the question ‘who is there and what are they doing?’. First, the GeoChip is a DNA microarray containing oligonucleotide probes for genes involved in biogeochemical cycling of carbon, nitrogen, phosphorus, sulfur and various metals, antibiotic resistance, organic contaminant degradation, virial activity, stress responses, and energy production, as well as gyrB-based phylogenetic markers. Thus, the GeoChip is able to specifically, sensitively, and quantitatively detect hundreds of thousands of microbial functional genes/groups important to biogeochemical, ecological and environmental processes. Second, GeoChip is a generic tool in its ability to analyze microbial samples from any environmental source, such as soil, water, air, and human and animal bodies. Third, GeoChip provides easy operation since it can analyze microbial samples without any culturing required, and without prior knowledge of a sample’s microbial composition. In addition, GeoChip is an unprecedented tool in its ability to rapidly and comprehensively identify the microbial community functional structure, activity and dynamics using both community DNA or RNA samples. Finally, one of the bottlenecks in using metagenomic tools for addressing environmental questions is the lack of appropriate standards for data comparison. The GeoChip allows comparable analysis of microarray data across different sites, experiments, laboratories and/or time periods by implementing universal standards.

GeoChips can be fabricated on a glass slide by spotting or in situ synthesis. Once DNA or RNA samples are obtained, numerous samples from one or more specific environments can be analyzed virtually on a daily basis. With the GeoChip pipeline, all results can be processed immediately after hybridization. Such rapid detection enables scientists to track functional processes over a short period of time, which was not previously possible.
Technology Features
Many studies have demonstrated that GeoChip is a powerful tool for specific, sensitive and quantitative detection of key functional genes from diverse microbial communities.
One of most important parameters to ensure the generation of high quality microarray data is specificity. This is especially true for analyzing environmental samples since there may be numerous sequences for each gene present in a sample. GeoChip specificity is controlled by probe design and hybridization conditions. Our studies showed that hybridizations at 50° C and 50% formamide could differentiate sequences with < 90-92% identity (Rhee et al., 2004; Liebich et al., 2006; Deng et al. 2008). With such high resolution, GeoChip is able to differentiate microorganisms at the species-strain level. Based on these results, we systematically tested and experimentally established probe design criteria by considering sequence identity, continuous sequence stretches and free energy in order to increase specificity (He et al., 2005; Liebich et al., 2006). These criteria have been implemented in a novel software tool, CommOligo, for microarray probe design (Li et al., 2005). Probes for GeoChips were designed using this newly developed software. For example, evaluation of GeoChip 2.0 revealed only a very small percentage of false positives (0.002-0.004%) and no false negatives under the experimental conditions examined (He et al., 2007). Thus, GeoChip has been extensively validated, and proven to be a highly specific microarray.
Sensitivity is another major concern, especially for samples from complex environments where many gene variants are expected to be in low abundance. GeoChips with 50mer oligonucleotide probes have shown that the detection limits were 5~10 ng of genomic DNA in the absence of background DNA, 50~100 ng of pure-culture genomic DNA in the presence of background DNA, and ~107 cells (Rhee et al., 2004; Tiquia et al., 2004). Such detection sensitivity is sufficient for analyzing environmental samples where microbial populations are abundant such as those from bioreactors and waste water treatment systems. But it is not high enough for analyzing most environmental samples in natural settings such as soils, groundwater, and marine water columns because the population abundance in these samples is generally lower than the detection limits. As a result, this will limit application of microarrays for many environmental studies.
To attack such challenges, we developed a novel, whole community genome amplification (WCGA)-assisted microarray detection approach for representative, sensitive, and quantitative detection and analysis of microbial communities whose members could not be studied using conventional microarray technology. Representative detection of individual genes or genomes was obtained with 1 to 500 ng community DNA. Lower concentrations of DNA (down to 10 fg, 2 bacterial cells, Wu et al. 2006) could be detected using a modified amplification buffer. Application of this approach to microbial communities in contaminated groundwaters demonstrated that it is feasible to biostimulate the indigenous microbial populations for contaminant remediation. This was the first demonstration that microarrays can be used in a high-throughput fashion to visualize the functional structure of microbial communities in natural settings with low biomass.
While DNA detection provides information on the presence of functional genes in the environment, it does not provide information on microbial activity. To monitor microbial activity RNA must be detected. However, while WCGA is able to amplify DNA, it cannot be used for RNA-based activity analyses. Thus, another approach, termed whole community RNA amplification (WCRA), was developed to provide sufficient amounts of mRNA from environmental samples for microarray analysis (Gao et al. 2007). Very representative detections were obtained with RNA of 50-100 ng, as low as with 10 ng of RNA, which is sufficient for detecting active populations in many natural settings. The developed method was successfully applied to active microbial populations in a fluidized bed reactor used for denitrification of contaminated groundwaters and to ethanol stimulated groundwater samples used for uranium reduction. This is the first demonstration that microarray-based technology can be used to successfully detect the activities of microbial communities from real environmental samples in a high-throughput fashion.
Another important issue is whether GeoChip can provide quantitative information. We have demonstrated that linear relationships were observed between target DNA or RNA concentrations and hybridization signal intensity using pure culture, mixed culture, and environmental samples without amplification (Rhee et al., 2004; Wu et al., 2001; Wu et al., 2006). With the WCGA approach, robust quantitative detection was observed by significant linear relationships between signal intensity and initial DNA concentrations or cell numbers (Wu et al. 2006). These studies demonstrate that GeoChip can be used to quantitatively analyze environmental samples.
Another important issue is whether GeoChip can provide quantitative information. We have demonstrated that linear relationships were observed between target DNA or RNA concentrations and hybridization signal intensity using pure culture, mixed culture, and environmental samples without amplification (Rhee et al., 2004; Wu et al., 2001; Wu et al., 2006). With the WCGA approach, robust quantitative detection was observed by significant linear relationships between signal intensity and initial DNA concentrations or cell numbers (Wu et al. 2006). These studies demonstrate that GeoChip can be used to quantitatively analyze environmental samples.
History & Legacy
The first functional gene array was constructed with 763 genes involved in nitrogen cycling (nirS, nirK, nifH, amoA), methane oxidation (pmoA), and sulfite reduction (dsrAB) (Tiquia et al., 2004). Then, a similar expanded array was developed with 2,402 genes involved in organic contaminant biodegradation and metal resistance (Rhee et al., 2004). Those arrays were referred to as GeoChip 1.0.
GeoChip 2.0 contained 24,243 oligonucleotide (50 mer) probes and covered more than 10,000 genes in 150 functional gene families involved in nitrogen, carbon, sulfur and phosphorus cycling, metal reduction and resistance, and organic contaminant degradation (He et al., 2007).
GeoChip 3.0 contained about 28,000 probes covering approximately 57,000 gene variants from 292 functional gene families involved in carbon, nitrogen, phosphorus and sulfur cycles, energy metabolism, antibiotic resistance, metal resistance and organic contaminant degradation. GeoChip 3.0 also had several other distinct features, such as a common oligo reference standard (CORS) for data normalization and comparison (Liang et al., 2010), a software package for data management and future updating and the gyrB gene for phylogenetic analysis (He et al., 2010).
GeoChip 4.0 contained approximately 82,000 probes covering 141,995 coding sequences from 410 functional gene families related to microbial carbon, nitrogen, sulfur, and phosphorus cycling, energy metabolism, antibiotic resistance, metal resistance/reduction, organic remediation, stress responses, bacteriophage, and virulence. As the last version, GeoChip 4.0 was developed on a NimbleGen 12x135K array platform that each chip contained 12 arrays (Tu et al., 2013, submitted).
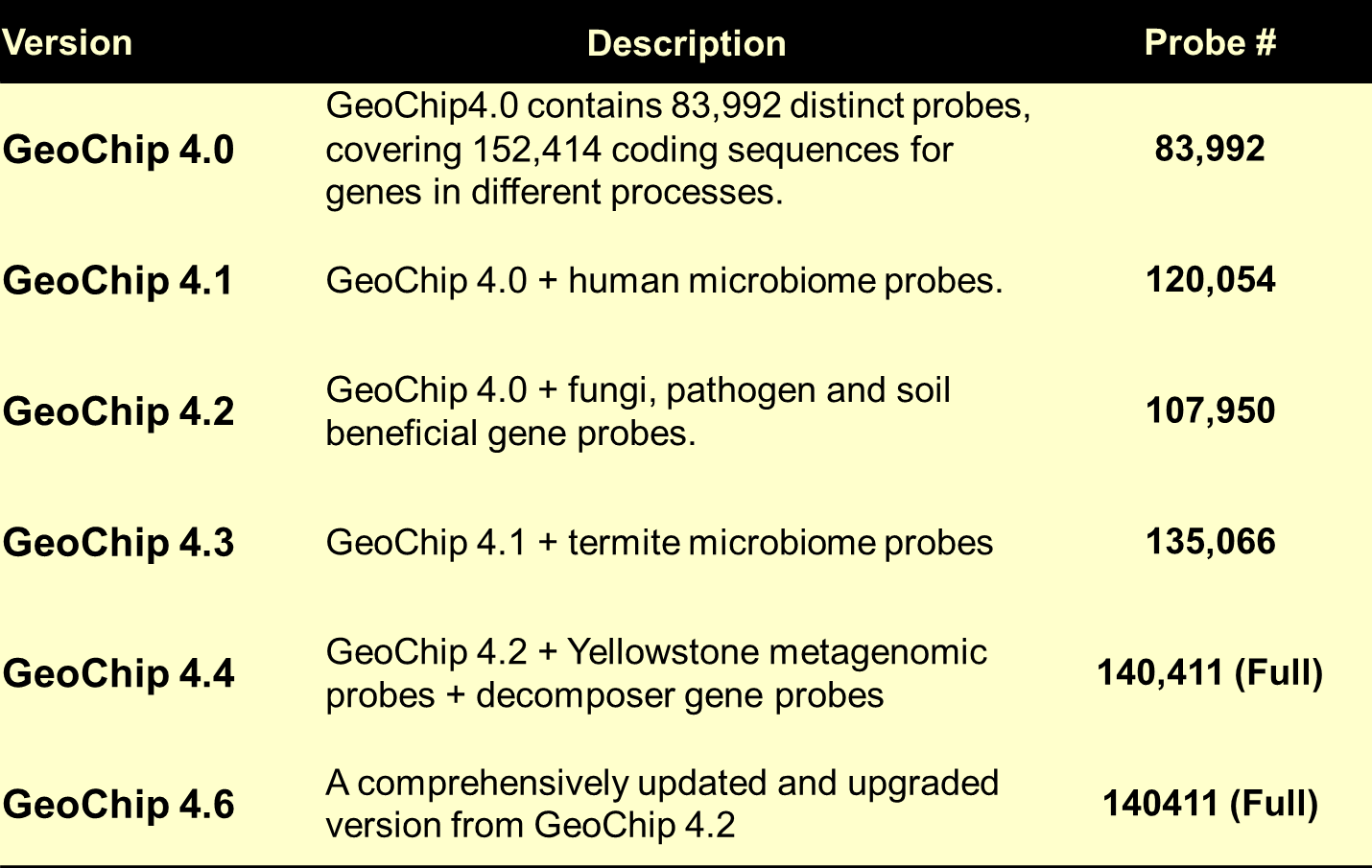
GeoChip 5.0
GeoChip 5.0 is the current version in use. GeoChip 5.0 contains 167,044 distinct probes, covering 395,894 coding sequences (CDS) from ~1500 functional gene families involved in microbial carbon (degradation, fixation, methane), nitrogen, sulfur, and phosphorus cycling, energy metabolism, metal homeostasis, organic remediation, “Other” (phylogenetic genes and CRISPR system), secondary metabolism (e.g. antibiotic metabolism, pigments), stress responses, viruses (both bacteriophages and eukaryotic viruses), and virulence. As a new array, GeoChip 5.0 was developed on the Agilent platform with Small (60 K x 8, 8 arrays with 60,000 probes each on one slide), Medium (180 K x 4), Large (400K x2) and Extra Large (1.0 M x 1) formats
GeoChip is a specific type of microarrays with each probe targeting a specific nucleic acid sequence or a group of highly similar sequences, and it can be constructed by spotting or in situ synthesis on a glass slide. DNA or RNA is extracted from a sample, such as soil, water, or food, and labeled with a fluorescent dye. When the labeled DNA or RNA molecules come into contact with the appropriate probe, the DNA is “captured” by the probe. After hybridization, free DNA/RNA molecules are washed away, and the signal intensity of the fluorescently labeled and captured DNA/RNA molecules is digitally imaged by a laser scanner. This digital image is then used to assess the concentration or abundance of the target DNA/RNA. GeoChip can simultaneously measure many gene sequences from one sample.
Several generations of GeoChips have been developed, which are classified as generic GeoChips since they contain genes involved in basic functional processes and from a variety of microorganisms, such as archaea, bacteria, and fungi. Such arrays are called Pan-GeoChips to distinguish them from specific microbiome chips, which target specific microbial communities, populations, functional processes, or environments. Currently, specific microarrays, such as FungChip, PathoChip and StressChip have been developed, and they are available for microbial community analysis.
FungChip
Fungal chip (FungChip) is mainly designed to detect and characterize fungal communities from different habitats or ecosystems.
StressChip
Microbial community responses to environmental stresses are critical for microbial growth, survival, and adaptation. To fill major gaps in our ability to discern the influence of environmental changes on microbial communities from engineered and natural environments, a functional gene based microarray, termed StressChip has been developed. The StressChip contains a total of 22,855 probes covering 79,628 coding sequences from 46 key functional genes involved in microbial responses to environmental stresses, such as temperature, osmolarity, oxidative status, nutrient limitation, and general stress response, which are derived from 46 key 985 bacterial, 76 archaeal, and 59 eukaryotic species/strains. The StressChip provides a new tool for accessing microbial community functional structure and responses to environmental changes (Zhou et al., 2013, Environmental Science and Technology).
PathoChip
A pathogen chip (PathoChip) is designed to assess pathogenic properties of diverse microbial communities in their environments and constructed with key virulence genes related to major virulence factors, such as adherence, colonization, motility, invasion, toxin, immune evasion and iron uptake. The Pathogen contains a total of 3,715 probes from 13 virulence factors, covering 7,417 coding sequences from 1,397 microbial species (2,336 strains), providing a useful tool to identify virulence genes in microbial populations, examine the dynamics of virulence genes in response to environmental perturbations and determine the pathogenic potential of microbial communities (Lee et al., the ISME Journal).
